Research
I am a Bioinformatics Fellow at the Gladstone Institues advised by Dr. Barbara Engelhardt and a part-time consultant for NE47 Bio. My expertise is in Bayesian statistical modeling for biological data. I currently work primarily on machine learning for experimental protein phenotype prediction and engineering and computer vision methods for phenotyping imaging time-series.
Postdoctoral Work - Gladstone Institutes
Stay tuned!
PhD Work - Princeton CBE
Published Work
A self-exciting point process to study multicellular spatial signaling patterns
with Siddhartha Jena
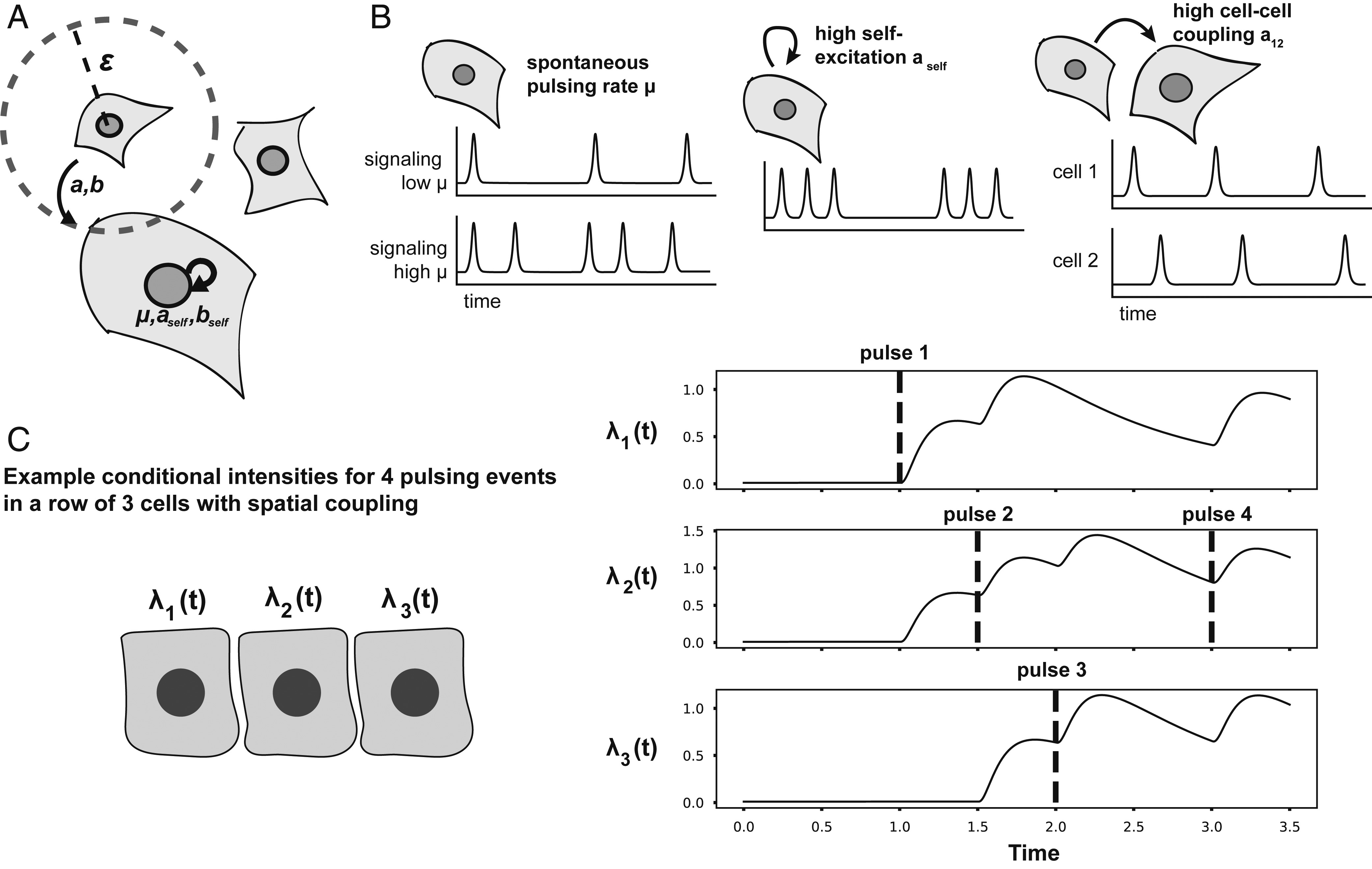
Cells are under constant pressure to integrate information from both their environment and internal cellular processes. However, these effects often use the same signaling pathways, making autonomous and coupled signaling difficult to decouple from one another. We present a statistical modeling framework, the cellular point process (CPP), that decouples these two modes of signaling using videos of living, actively signaling cells as input. Our model reveals modulation of autonomous and coupled signaling parameters in a number of contexts ranging from pharmacological treatment to wound healing that were previously unavailable. The CPP enhances our understanding of cellular information processing and can be extended to a wide range of systems. in PNAS
A robust nonlinear low-dimensional manifold for single cell RNA-seq data
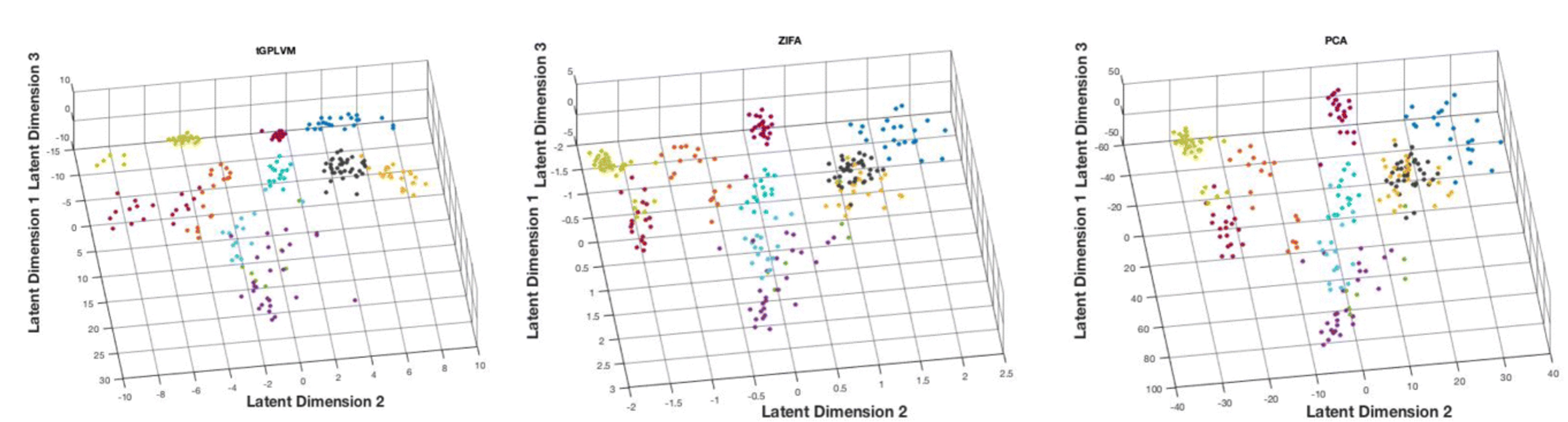
Modern developments in single-cell sequencing technologies enable broad insights into cellular state. Single-cell RNA sequencing (scRNA-seq) can be used to explore cell types, states, and developmental trajectories to broaden our understanding of cellular heterogeneity in tissues and organs. Analysis of these sparse, high-dimensional experimental results requires dimension reduction. Several methods have been developed to estimate low-dimensional embeddings for filtered and normalized single-cell data. However, methods have yet to be developed for unfiltered and unnormalized count data that estimate uncertainty in the low-dimensional space. We present a nonlinear latent variable model with robust, heavy-tailed error and adaptive kernel learning to estimate low-dimensional nonlinear structure in scRNA-seq data. in BMC Bioinformatics
Preprints
A Bayesian nonparametric semi-supervised model for integration of multiple single-cell experiments
Joint analysis of multiple single cell RNA-sequencing (scRNAseq) data is confounded by technical batch effects across experiments, biological or environmental variability across cells, and different capture processes across sequencing platforms. Manifold alignment is a principled, effective tool for integrating multiple data sets and controlling for confounding factors. We demonstrate that the semi-supervised t-distributed Gaussian process latent variable model (sstGPLVM), which projects the data onto a mixture of fixed and latent dimensions, can learn a unified low-dimensional embedding for multiple single cell experiments with minimal assumptions. We show the efficacy of the model as compared with state-of-the-art methods for single cell data integration on simulated data, pancreas cells from four sequencing technologies, induced pluripotent stem cells from male and female donors, and mouse brain cells from both spatial seqFISH+ and traditional scRNA-seq. preprint
Nonparametric Deconvolution Models
with Allison Chaney
We describe nonparametric deconvolution models (NDMs), a family of Bayesian nonparametric models for collections of data in which each observation is the average over the features from heterogeneous particles. For example, these types of data are found in elections, where we observe precinct-level vote tallies (observations) of individual citizens’ votes (particles) across each of the candidates or ballot measures (features), where each voter is part of a specific voter cohort or demographic (factor). Like the hierarchical Dirichlet process, NDMs rely on two tiers of Dirichlet processes to explain the data with an unknown number of latent factors; each observation is modeled as a weighted average of these latent factors. Unlike existing models, NDMs recover how factor distributions vary locally for each observation. This uniquely allows NDMs both to deconvolve each observation into its constituent factors, and also to describe how the factor distributions specific to each observation vary across observations and deviate from the corresponding global factors. We present variational inference techniques for this family of models and study its performance on simulated data and voting data from California. We show that including local factors improves estimates of global factors and provides a novel scaffold for exploring data.
Other Work
Published work
A generative model for predicting terrorist incidents
A major concern in coalition peace-support operations is the incidence of terrorist activity. In this paper, we propose a generative model for the occurrence of the terrorist incidents, and illustrate that an increase in diversity, as measured by the number of different social groups to which that an individual belongs, is inversely correlated with the likelihood of a terrorist incident in the society. A generative model is one that can predict the likelihood of events in new contexts, as opposed to statistical models which are used to predict the future incidents based on the history of the incidents in an existing context. Generative models can be useful in planning for persistent Information Surveillance and Reconnaissance (ISR) since they allow an estimation of regions in the theater of operation where terrorist incidents may arise, and thus can be used to better allocate the assignment and deployment of ISR assets. In this paper, we present a taxonomy of terrorist incidents, identify factors related to occurrence of terrorist incidents, and provide a mathematical analysis calculating the likelihood of occurrence of terrorist incidents in three common real-life scenarios arising in peace-keeping operations. in SPIE
Addressing the Limitations of AI/ML in creating Cognitive Solutions
In order to solve real-world problems associated with development of Cognitive Solutions, several key problems need to be addressed. The current state of art in AI/ML is too limited for it to offer practical solutions that will outperform manually developed solutions in a large number of contexts. Building upon experience developed in creating AI/ML solutions for enterprise networks and IoT focused solutions, we will outline the limitations of current AI/ML systems, and discuss ways to address those challenges. We would also describe the vision of an AI/ML based system that ought to be developed in order to attain the vision of a true cognitive AI/ML systems that can be used in a broad set of enterprise contexts. in IEEE CogMI
Preprints
Differentially methylated regions and methylation QTLs for teen depression and early puberty in the Fragile Families Child Wellbeing Study
My research journey
I’ve been involved in academic research since high school, where I worked under the supervision of Albert Einstein College of Medicine with Joel Friedman and directly under Mahantesh Navati and Camille LaRoche. There I worked on the synthesis of hemoglobin-containing solution-gelatins and sugar glasses to test reactions with nitric oxides.
After high school, I followed this interest in biology with a Biomedical Engineering major at Duke University. I joined Ashutosh Chilkoti’s group and worked with Kelli Lubinguhl and Jayanta Bhattacharyya on a variety of projects cented on improving drug delivery with elastin-like polypeptides. Target drugs included the biological betatrophin for diabetes and paclitaxel and gemcitabine for cancer.
In my doctoral work I transitioned to a computational role, advised by Dr. Barbara Engelhardt in the Princeton Computer Science department. I combined my engineering experience with machine learning tools to answer questions about bulk RNA-seq, single cell RNA-seq, spatial transcriptomics, and live cell imaging with statistical methods.
After my completing my PhD in 2021 I joined IBM as a data scientist in the Federal Analytics and Cognitive Consulting division. My work there focused on detecting anomalies in ship tracking data and predicting medical supply shortages from time series data.
I returned to work with Dr. Engelhardt as a Bioinformatics Fellow in 2022 to return to my biology roots. I am currently working primarily on machine learning for proteins and biological imaging.